4.1. PTM Interaction Network Visualization
The Network View tab of the results page provides visualisation of all of your query protein's mutations and/or PTM sites that are known to be bound by protein kinases, as well as, which of those can be targeted by drugs (based on experimental data and MIMP prediction):
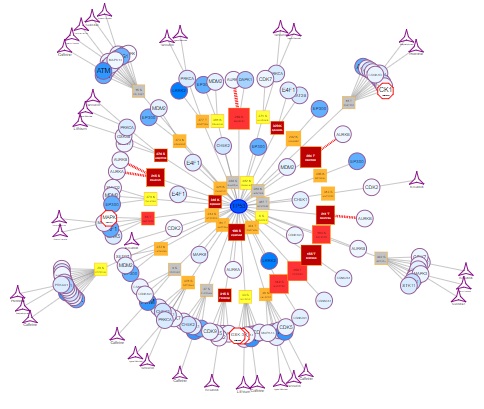
At the centre of the PTM interaction map is your query protein as depicted by an ellipse with a thick border.
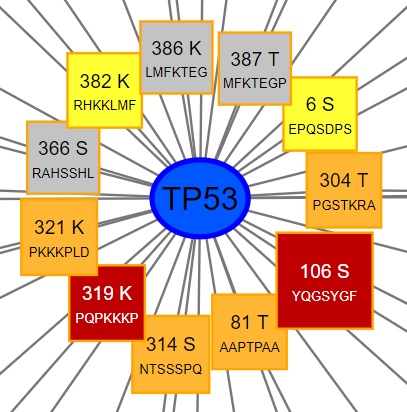
Surrounding (and connected to) the query protein are mutations affecting the protein's PTM sites. Each PTM site is visualised as a coloured box listing the mutation location and the surrounding amino acid sequence:


|
Mutation Impact |
Explanation |
|
Direct |
mutation affects the post-translationally modified amino acid |
|
Proximal |
mutation affects an amino acid located 1-3 amino acids away from the PTM site |
|
Distal |
mutation affects an amino acid located 4-7 amino acids away from the PTM site |
|
Rewiring |
mutation-induced gains and losses of kinase-bound sequence motifs (predicted by the MIMP software) |
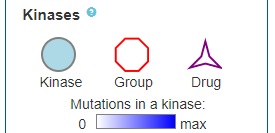
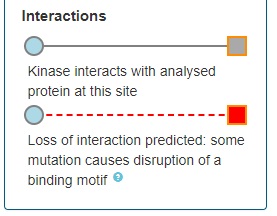
The mutations located in your protein of interest are further connected with kinases which are shown to bind the PTM. The colour of the circles corresponds to the number of mutations occurring in the kinase (white = no mutations; dark blue = most mutated). Furthermore, these kinases (circles) or kinase families (octagon) are connected the respective kinase-targeting drugs (stars):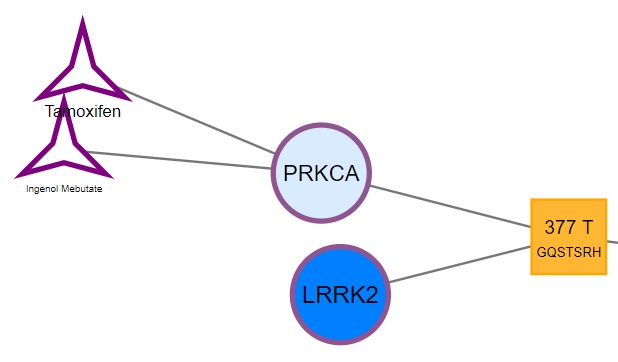
Zooming in/out
If you are interested in a specific PTM site, you can click on it to zoom onto it and the kinases it has been associated with. Alternatively you can click the magnifying glass icons on the top right of the PTM Interaction Network Visualisation window. Lastly, you can also use your mouse scroll wheel to zoom in and out of the image.
Saving the image
The Interaction Network image can be saved in an SVG or PDF formats by clicking on the “Save image as” button on the top right of the PTM Interaction Network Visualisation window.