ActiveDriverDB'20
Find your mutation or gene of interest


What is ActiveDriverDB?
ActiveDriverDB is a human proteo-genomics database. The database uses information about post-translational modifications (PTMs) in proteins to annotate and interpret genetic variation, disease genes, and cancer driver genes. All datasets in ActiveDriverDB is collected from public resources and based on experimental data. The datasets can be explored starting from some example queries (dementia mutations in MAPT, cystic fibrosis mutations in CFTR, cancer mutations in CTNNB1 phosphodegron), browsed interactively, downloaded in bulk or retrieved using an application program interface (API). Users can also upload their own custom datasets and store on the server for a limited time.
What's new?
This is a 2020 version of the ActiveDriverDB. Please see our changelog. To access the previous (2017) version click here. To access the newer (2021) version click here.
Example
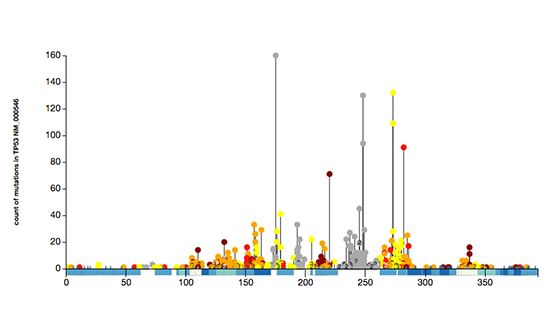
Sequence
Network
About the data
Post-translational modifications (PTMs) are chemical modifications of amino acids that act as molecular switches and expand the functional repertoire of proteins. Genetic changes in the protein sequence, such as mis-sense single nucleotide variants (SNVs) collected in this database, often change the sequence and structure of sites corresponding to PTMs. Further, PTMs are created and erased by certain enzymes, such as kinases that cause protein phosphorylation. These enzymes recognize specific motifs in protein sequence that may also be changed by SNVs. Thus interpreting genetic variation with protein PTMs may reveal new mechanistic details of genetic variation and protein function and help decipher genotype-phenotype associations in human variation and disease.
You can browse gene and pathways, ranked by PTM mutations and/or FDR:
Proteins |
39,159 |
| Kinases and other PTM enzymes |
655 |
| Kinase families |
143 |
Mutations
|
3,707,271 |
| ClinVar |
318,428 |
| TCGA MC3 | 1,595,400 |
| ESP6500 | 1,318,972 |
| 1000 Genomes |
1,066,906 |
| Mutations in PTM sites |
826,828 |
|
Mutations network-rewiring effect |
70,734 |
| Total number of annotated nucleotides | 96,138,347 |
Pathways |
18,511 |
| Kinase - site interactions | 33,840 |
| Kinases interacting with >= 1 site | 497 |
PTM sites |
544,720 |